CAN3D: Fast 3D medical image segmentation via compact context aggregation |
| |
| Affiliation: | 1. School of Automation, Northwestern Polytechnical University, Xi''an, 710072, China;2. Center for Brain and Brain-Inspired Computing Research, School of Computer Science, Northwestern Polytechnical University, Xi''an 710129, China;3. Cortical Architecture Imaging and Discovery Lab, Department of Computer Science and Bioimaging Research Center, The University of Georgia, Athens, GA, USA;1. Institute for Biomedical Engineering, University and ETH Zurich, Zurich, Switzerland;2. Laboratoire de Mécanique des Solides (LMS), École Polytechnique/C.N.R.S./Institut Polytechnique de Paris, Palaiseau, France;3. MΞDISIM team, Inria, Palaiseau, France;1. Medical Image Processing Group, 602 Goddard building, 3710 Hamilton Walk, Department of Radiology, University of Pennsylvania, Philadelphia, PA 19104, United States;2. Quantitative Radiology Solutions, LLC, 3675 Market Street, Suite 200, Philadelphia, PA 19104, United States |
| |
| Abstract: | 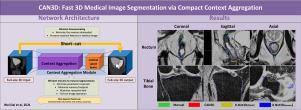
Direct automatic segmentation of objects in 3D medical imaging, such as magnetic resonance (MR) imaging, is challenging as it often involves accurately identifying multiple individual structures with complex geometries within a large volume under investigation. Most deep learning approaches address these challenges by enhancing their learning capability through a substantial increase in trainable parameters within their models. An increased model complexity will incur high computational costs and large memory requirements unsuitable for real-time implementation on standard clinical workstations, as clinical imaging systems typically have low-end computer hardware with limited memory and CPU resources only. This paper presents a compact convolutional neural network (CAN3D) designed specifically for clinical workstations and allows the segmentation of large 3D Magnetic Resonance (MR) images in real-time. The proposed CAN3D has a shallow memory footprint to reduce the number of model parameters and computer memory required for state-of-the-art performance and maintain data integrity by directly processing large full-size 3D image input volumes with no patches required. The proposed architecture significantly reduces computational costs, especially for inference using the CPU. We also develop a novel loss function with extra shape constraints to improve segmentation accuracy for imbalanced classes in 3D MR images. Compared to state-of-the-art approaches (U-Net3D, improved U-Net3D and V-Net), CAN3D reduced the number of parameters up to two orders of magnitude and achieved much faster inference, up to 5 times when predicting with a standard commercial CPU (instead of GPU). For the open-access OAI-ZIB knee MR dataset, in comparison with manual segmentation, CAN3D achieved Dice coefficient values of (mean = 0.87 ± 0.02 and 0.85 ± 0.04) with mean surface distance errors (mean = 0.36 ± 0.32 mm and 0.29 ± 0.10 mm) for imbalanced classes such as (femoral and tibial) cartilage volumes respectively when training volume-wise under only 12G video memory. Similarly, CAN3D demonstrated high accuracy and efficiency on a pelvis 3D MR imaging dataset for prostate cancer consisting of 211 examinations with expert manual semantic labels (bladder, body, bone, rectum, prostate) now released publicly for scientific use as part of this work. |
| |
| Keywords: | Semantic segmentation Magnetic resonance imaging Deep learning Prostate and knee 3D |
| 本文献已被 ScienceDirect 等数据库收录! |
|

