Integrated pathway-based transcription regulation network mining and visualization based on gene expression profiles |
| |
| Affiliation: | 1. Graduate School of Information Science, Nara Institute of Science and Technology, Takayama 8916-5, Ikoma, Nara 630-0192, Japan;2. Department of Respiratory Medicine, Graduate School of Medicine, University of Tokyo, 7-3-1, Hongo, Bunkyo-ku, Tokyo 113-0033, Japan |
| |
| Abstract: | 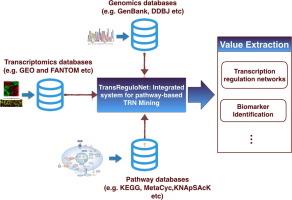
Conventionally, workflows examining transcription regulation networks from gene expression data involve distinct analytical steps. There is a need for pipelines that unify data mining and inference deduction into a singular framework to enhance interpretation and hypotheses generation. We propose a workflow that merges network construction with gene expression data mining focusing on regulation processes in the context of transcription factor driven gene regulation. The pipeline implements pathway-based modularization of expression profiles into functional units to improve biological interpretation. The integrated workflow was implemented as a web application software (TransReguloNet) with functions that enable pathway visualization and comparison of transcription factor activity between sample conditions defined in the experimental design. The pipeline merges differential expression, network construction, pathway-based abstraction, clustering and visualization. The framework was applied in analysis of actual expression datasets related to lung, breast and prostrate cancer. |
| |
| Keywords: | Biological interpretation Pathway-based modularization Gene regulation Transcription factors |
| 本文献已被 ScienceDirect 等数据库收录! |
|

