Development of a multilocus sequence typing tool for high-resolution subtyping and genetic structure characterization of Cryptosporidium ubiquitum |
| |
| Affiliation: | 1. Division of Foodborne, Waterborne and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA 30329, USA;2. College of Veterinary Medicine, Northeast Agricultural University, Harbin, Heilongjiang 150030, China;3. School of Resources and Environmental Engineering, East China University of Science and Technology, Shanghai 200237, China;1. College of Animal Science and Veterinary Medicine, Henan Agricultural University, 95 Wenhua Road, Zhengzhou, Henan 450002, PR China;2. College of Animal Science, Tarim University, Tarim Road 1487, Alar, Xinjiang 843300, PR China;3. Department of Animal Science, Henan Vocational College of Agriculture, Zhongmu Henan 451450, PR China |
| |
| Abstract: | 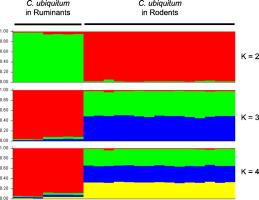
Cryptosporidium ubiquitum is an emerging zoonotic pathogen in humans. Recently, a subtyping tool targeting the 60-kDa glycoprotein (gp60) gene was developed for C. ubiquitum, and identified six subtype families (XIIa-XIIf). In this study, we selected five genetic loci known to be polymorphic in C. hominis and C. parvum for the development of a multilocus subtyping tool for C. ubiquitum, including CP47 (cgd6_1590), MSC6-5 (cgd6_4290), cgd6_60, cgd2_3690, and cgd4_370. PCR primers for these targets were designed based on whole genome sequence data from C. ubiquitum. DNA sequence analyses of 24 C. ubiquitum specimens showed the presence of 18, 1, 5, 4, and 5 subtypes at the CP47, MSC6-5, cgd6_60, cgd2_3690, and cgd4_370 loci, respectively. Altogether, 18 multilocus sequence typing (MLST) subtypes were detected among the 19 specimens successfully sequenced at all polymorphic loci. Phylogenetic analyses of the MLST data indicated that the rodent subtype families of XIIe and XIIf were highly divergent from others, and the ruminant XIIa subtype family formed a monophyletic group genetically distant from other rodent subtype families XIIb, XIIc, and XIId. The latter showed no consistent grouping of specimens and formed one large cluster in phylogenetic analysis of concatenated multilocus sequences. This was supported by results of STRUCTURE and FST analyses, which further suggested that XIIa originated from one common ancestor whereas XIIb, XIIc, and XIId contained mixed ancestral types, reflecting a close relatedness of the three subtype families and the likely occurrence of genetic recombination among them. Thus, an MLST tool was developed for high-resolution subtyping of C. ubiquitum and results of preliminary characterizations of specimens from humans and animals supported the conclusion on the existence of ruminant and rodent-adapted C. ubiquitum groups. |
| |
| Keywords: | |
| 本文献已被 ScienceDirect 等数据库收录! |
|

